超级增强子研究
超级增强子简介
超级增强子 (Super Enhancer, SE) 为8-20 kb具有转录增强活性的超长顺式作用元件,可富集高密度的关键转录因子(master transcription factors)、辅因子(cofactor)和增强子表观修饰标记(histone modification marks),并激活细胞中身份决定基因的表达,发挥调控细胞命运的关键作用,还能促进关键致癌基因表达,富集疾病相关变异,具有重大研究价值,已经成为生物医学领域的研究热点。
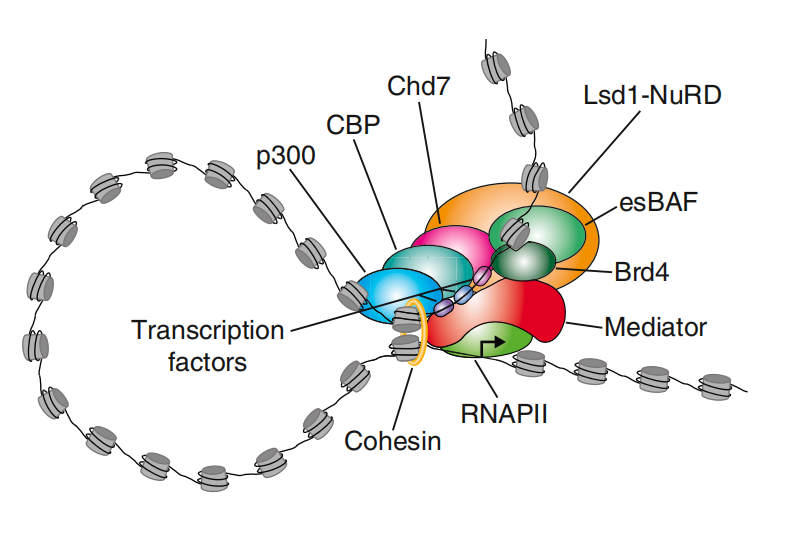
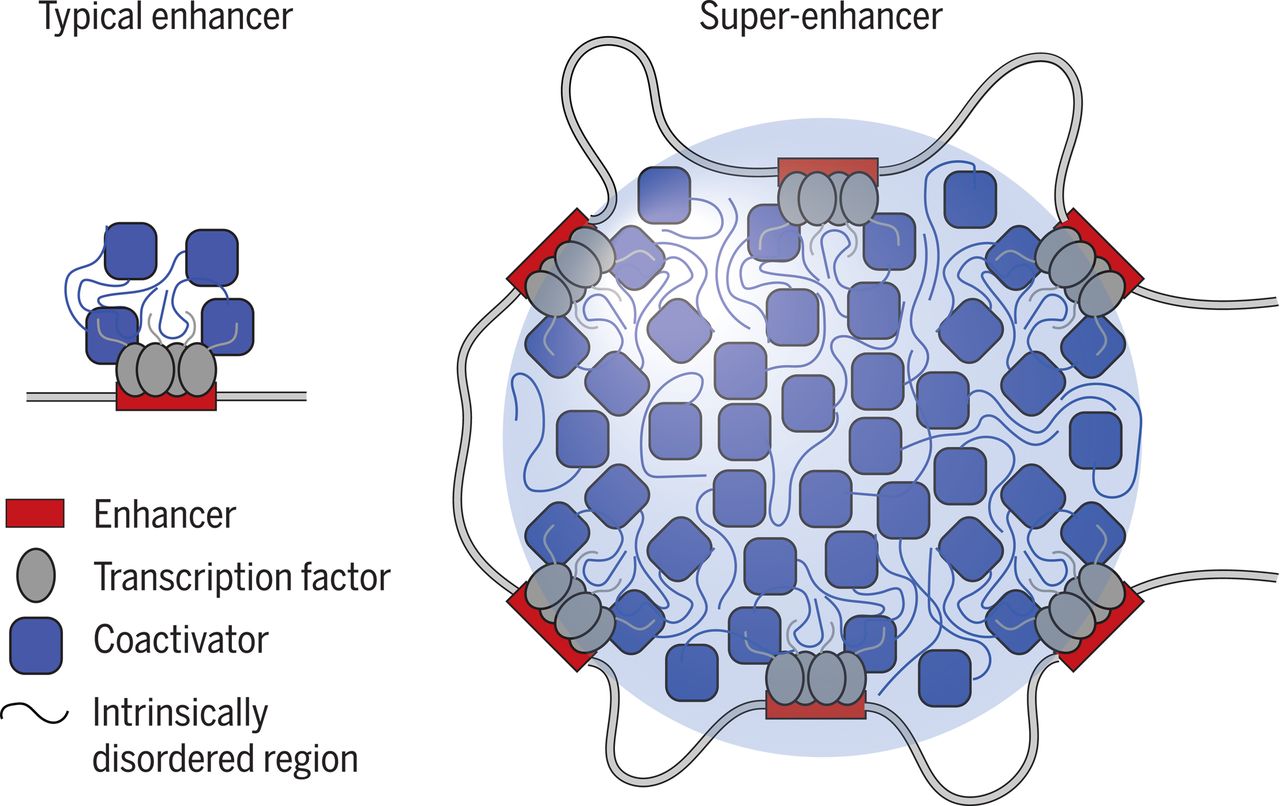
超级增强子的研究意义
1、发现致病驱动基因;
2、分析疾病关联变异位点易感性;
3、开发基于SEs复杂性疾病的精准诊断和药物开发
超级增强子的鉴定
目前对增强子鉴定,主要采用染色质免疫共沉淀技术(ChIP-seq)针对活性增强子相关联的因子或组蛋白修饰进行检测,如转录因子、转录辅激活因子(如Mediator、p300)、组蛋白修饰H3K27ac 和H3K4me1等。活性增强子通常同时含有H3K27ac 和H3K4me1修饰,而静态增强子(poised enhancer)一般同时具有H3K4me1和H3K27me3组蛋白标记。在此基础上,超级增强子依据增强子转录活性标记分子结合水平强度的差异进行鉴定。在分析方法上,首先对所得增强子进行缝合。主要依据在基因组范围内,这些单个增强子实体间如在12.5 kb 范围内,则合并为单个实体,即缝合增强子(Stitched enhancer)。最后,确定超级增强子和普通增强子之间的阈值。缝合增强子和其余的单个增强子按照ChIP-seq所测信号水平的强度排序,绘制获得一张曲线图,该曲线上斜率为1 的切线的切点所得的信号值为区分超强增强子和普通增强子之间的阈值,高于该值为超级增强子,其余的则称为普通增强子(Typical enhancer)。
超级增强子的鉴定流程
超级增强子研究设计的技术
A、ChIP-seq/CUT & Tag
通过染色质免疫共沉淀技术(ChIP-seq)或者CUT & Tag技术,针对活性增强子相关联的因子或组蛋白修饰进行检测,再利用ROSE算法,鉴定出超强增强子和普通增强子。
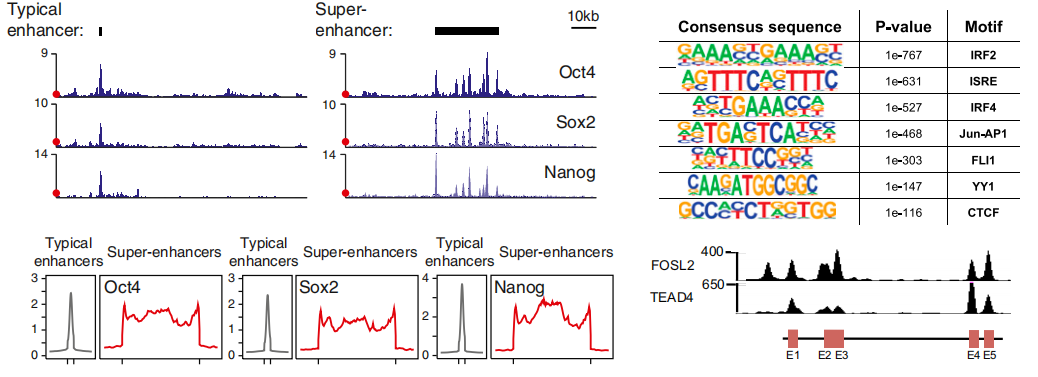
SE与TE信号图、motif分析
超级增强子的注释及调控靶基因预测
B、ATAC-seq
ATAC-seq(Assay for Transposase-Accessible Chromatin with highthroughput sequencing)利用 Tn5 酶可以进入细胞核并切割暴露的 DNA,并且 Tn5 酶在切割的同时可以在 DNA 的两端连上已知 DNA 序列标签,利用已知 DNA 序列的标签进行 PCR 扩增后测序,就可以识别出染色质开放区域,从而捕获调控序列的信息。
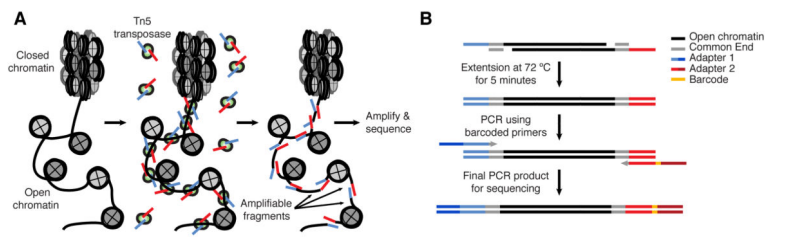
ATAC原理及建库流程
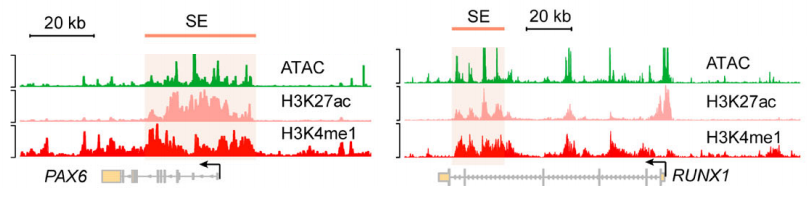
通过ATAC-seq Peak 数据与SE数据取交集,排除部分处于关闭转态的SE。
C、超级增强子功能缺失性研究
1.整体抑制:JQ1(Brd4抑制剂)
JQ1是BET蛋白的小分子抑制剂,可与BET溴结构域乙酰化赖氨酸残基竞争性结合,从而阻断含溴结构域的蛋白质(BRD)的功能。
2. 靶向抑制:EpiTM dCas9-NLS-KRAB
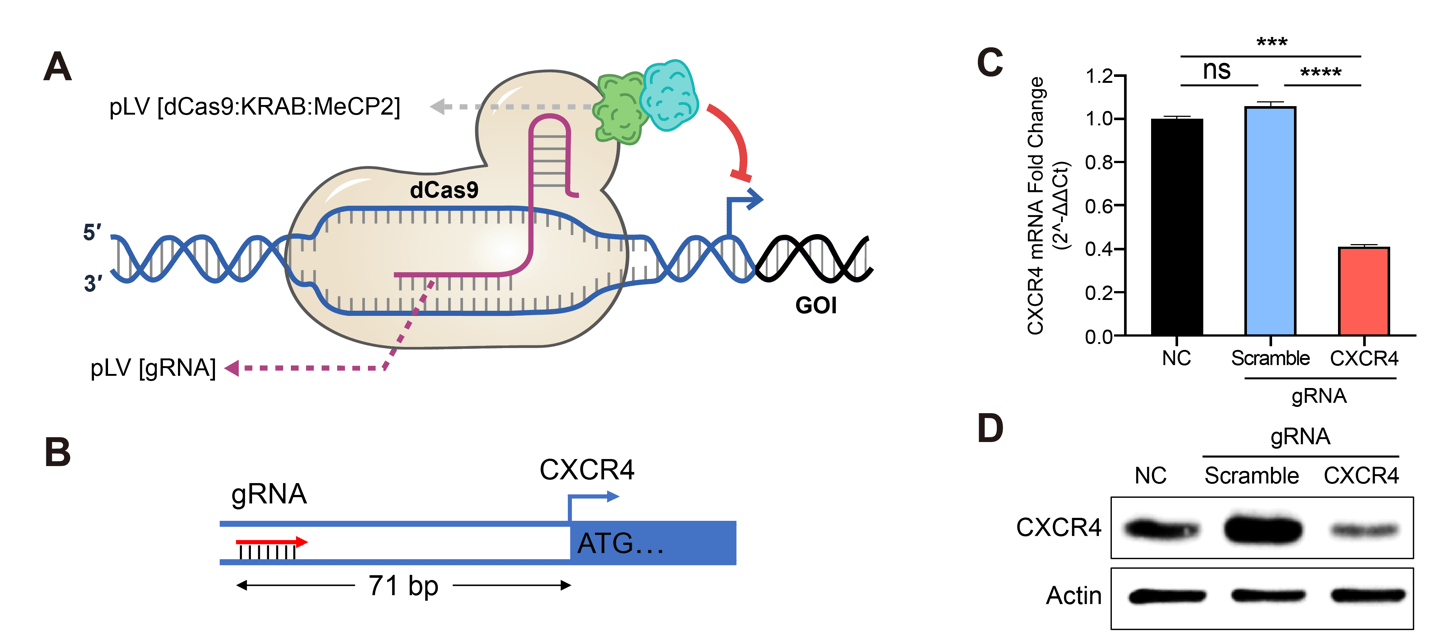
D、超级增强子功能获得性研究
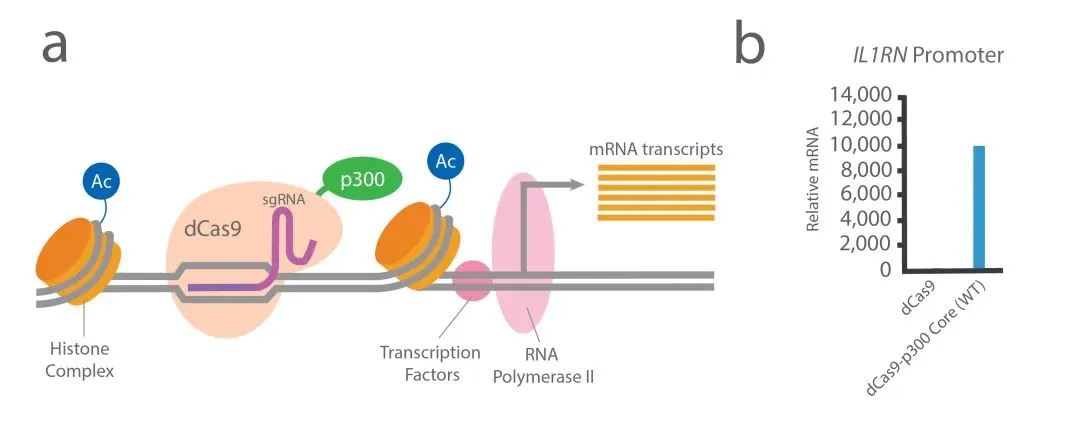
E、Crispr/Cas9
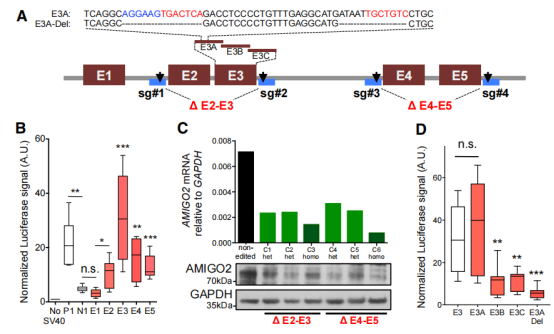
F、Luciferase assays